Dynamics of chromatin architecture in the context of gene transcription
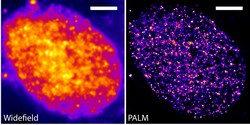
DNA is wrapped around a core histone structure to form nucleosomes, which are then folded further to generate chromatin fibres and then chromosomes. Chromatin organisation and compaction state change during activation or repression of gene transcription, a process accompanied by gene repositioning inside the nucleus. Nevertheless, the regulatory role played by the chromatin architecture during the modulation of transcription remains elusive. Scientists on the EU-funded CHROMATRANSCRIPT (Role of the chromatin architecture in the regulation of gene transcription) project set out to investigate aspects of the nuclear restructuring process in association with modulation of gene transcription. In this context, researchers developed methodologies for the quantitative characterisation of the dynamics of chromatin architecture. They then studied gene transcription using the oestrogen-regulated genes as a model system. The generated technique utilised fluorophore-conjugated histones that allowed studying the remodelling of specific sub-nuclear regions. In an alternative approach, fluorescent DNA was used to assess the local chromatin movements from milliseconds up to minutes. Furthermore, researchers succeeded at analysing the dynamics of single fluorescently labelled nucleosomes in living cells. This allowed them to characterise nucleosome motion at an unprecedented spatio-temporal resolution. Implementation of these techniques in human breast cancer cells showed a constrained diffusive motion of chromatin and a flexibility of chromatin fibres in living cells. Interestingly, estradiol-induced gene transcription did not affect chromatin steady-state dynamics. At DNA lesions, scientists observed a rapid chromatin relaxation triggered by the poly-ADP-ribosylation signalling pathway. Overall, project findings provide unprecedented resolution on chromatin movement in living cells and bring us a step closer to unravelling the mechanisms of chromatin dynamics. At the same time, the CHROMATRANSCRIPT tools will facilitate future research in the field of chromatin remodelling.