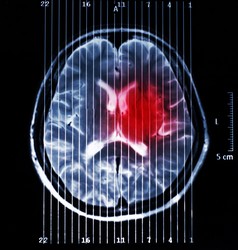
Bayesian methods to characterise brain tumours
Magnetic resonance imaging (MRI) is currently used for in vivo diagnosis and grading of brain tumours. This aids surgeons when planning tumour removal as well as in monitoring treatment response. However, diagnostic accuracy is still insufficient in some cases, for instance in patients who have malignant gliomas. It is mainly in these cases where magnetic resonance spectroscopy (MRS) and spectroscopic imaging (MRSI) play an important role. MR spectra (from MRS and MRSI) provide invaluable biochemical information through which precise metabolomic signatures can be obtained. As this information is very rich in content and most of the time difficult to interpret by doctors, we rely on pattern recognition techniques, which have proven successful at helping to improve diagnostic accuracy. However, there is still room for improvement of current pattern recognition methods so that they can assist with more complex cases and provide better results. Scientists of the EU-funded BAYESIANMULTIMODALMR (Multimodal analysis of MR data using Bayesian methods) project proposed a solution that uses pattern recognition techniques to integrate data obtained from MRI into MRSI. More specifically, they further improved the non-negative matrix factorisation (NMF) approach for tumour delineation in neuro-oncology by using as prior knowledge anatomical information of the tumour provided by the MRI. The methodology developed by the team for brain tumour segmentation allows for individual analysis of affected patients, which means that it avoids contamination from inter-subject overlaps. Also, it allows using only partial regions so that areas of uncertainty are excluded during initial segmentation. Testing with data from brain tumour-bearing mice demonstrated the feasibility of embedding MRI information into the source extraction in MRSI data. Researchers also developed a Bayesian NMF-based approach to automatically select relevant sources using the greedy strategy. This strategy helps remove redundant and duplicate data. Simulated and real-world data compiled by the INTERPRET European project were used for testing and validation. Another key development was a method for extracting meaningful source signals that can be used for tumour classification. These signals refer to healthy tissue, brain tumour response to therapy, and untreated or refractory brain tumours. The generated nosologic images proved the utility of this approach for disease classification and determination of tumour response to therapy. Research outcomes were disseminated widely at leading conferences and in international journals, amounting to about 13 peer-reviewed publications/communications. Implementation of the methods developed in this study would permit the integration of data from diverse imaging modalities for effective disease diagnosis, brain tumour grading and patient monitoring. This approach could also be adapted for the diagnosis of other difficult-to-detect neurological conditions.