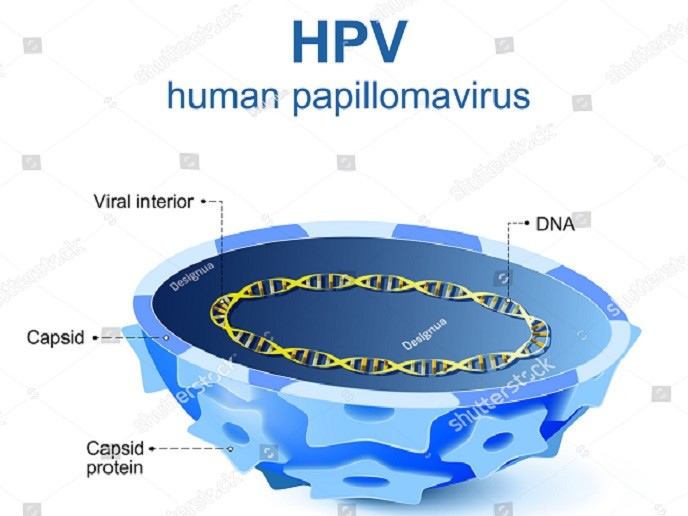
Papillomavirus evolution – the fight against cancer
The malignant transformation of certain PVs has been linked to the E5, E6 and E7 oncogenes present in the viral DNA genome. Although the proteins expressed by these genes have been extensively studied, there is little information on their evolutionary origin.
Studying the differences between oncogenic and non-oncogenic PVs
With the support of the Marie Skłowdowska-Curie programme, they combined computational and experimental methods to study specific events that occurred during PV genome evolution. As project coordinator Ignacio Bravo explains: “We applied the power of evolutionary thinking: we studied the genomes of extant PVs and the interactions between viral and human proteins, and reconstructed the evolutionary history of these genomes and these interactions.″ Researchers analysed the actual sequences of all genes in all known PVs, and through bioinformatics they inferred their history. They reconstructed their evolutionary relationships as a phylogenetic tree to determine if the ancestral PV genome contained all the genes present in the modern assembly, or whether they had been sequentially acquired. Interestingly, scientists discovered that the genome of the ancestral PVs some 400 million years ago only contained two gene cassettes: one for replication and one for producing virions. The E6 and E7 viral oncogenes emerged much later while the E5 oncogene seems to have appeared de novo from a DNA sequence some 40 million years ago. “Such appearance of new genes from scratch is one of the main sources of evolutionary novelty, enabling PVs to change the mode of exploiting the host,″ continues Bravo. From a single viral lineage, three types of PVs emerged: one causing hand warts, a second one causing genital warts, and a third one causing mucosal lesions that can lead to cancers. To investigate the oncogenic potential of the ancestral PV oncogenes, researchers ‘resurrected’ them experimentally through synthetic biology. Introduction into different human cell lines in culture enabled scientists to assess the impact of these oncoproteins on cellular fitness and further examine their interaction with cellular tumour suppressor proteins that could eventually lead to cancer.
Taking ONCOGENEVOL findings further
Research on PV-associated cancers has progressed enormously in the last thirty years. Although most sexually active humans become infected by PVs, only a small fraction will eventually develop cancer. Despite the identified genetic differences between oncogenic and non-oncogenic PVs, there is limited understanding on the determinant factors that render PVs oncogenic. The ONCOGENEVOL project expanded current knowledge on the origin of PV genes and genomes, by applying evolutionary medicine thinking to understand the long-term interplay between very common viruses and the cancers they cause. The project set the foundation to understand how certain PVs evolved their oncogenic potential. As the Marie Skłowdowska-Curie research fellow Anouk Willemsen explains “we have significantly improved our understanding of the evolutionary history of PVs and our results have great potential of being exploited for future development of PV vaccines.″ Future plans include the identification of the determinant factors of PV-induced cancer onset. This will be instrumental when designing public health interventions (including screening and vaccination) to reduce oncogenic PV infection and hence cancer incidence.