Tracing the early steps of HIV infection via protein signatures
Viruses are the ultimate parasites! A virus has very little genetic material, RNA in the case of HIV. So once inside the cell, it hijacks the host resources to produce its own proteins to make more viruses. Researchers with the hRBP-virion(opens in new window) project have developed new methods to dig out cellular proteins, known as RNA-binding proteins (hRBPs), that interact with viral RNAs. “Our aim is to identify ‘all’ the human RNA-binding proteins that are involved in virus infection by combining a new generation of proteomic, computational and functional analyses,” outlines Alfredo Castello Palomares, group leader at the coordinating institute, University of Oxford(opens in new window). Out of almost 1 600 hRBPs, 472 have already been linked to virus infection, based on our computational analysis of previous literature. To discover the RBPs involved in infection, the hRBP-virion team has developed a new experimental approach, RNA interactome capture (RIC), that was set up in a ‘safe’ viral system, sindbis(opens in new window). This cutting-edge method uncovered almost 250 RBPs that are altered in infected cells. “This tells us how the virus is regulating the cells, as many of these proteins are critical either for replication or to counter infection,” Castello points out. The Marie Skłodowska-Curie fellow on the research team, Manuel Garcia-Moreno, continues the story: “Once we had established the system in sindbis, we extended it to a critical human pathogen, HIV.”
Fishing for hRBPs, a question of using the right bait
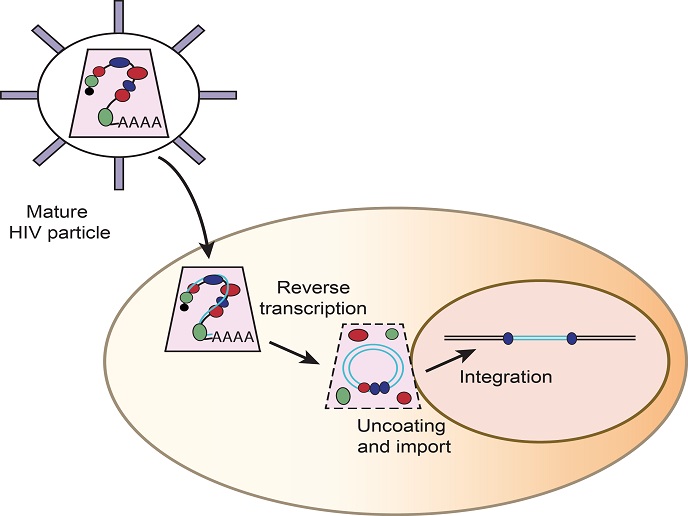
New research has reported that reverse transcription to make DNA from RNA happens inside the capsid shell of HIV. The HIV DNA is then integrated into the host cell DNA, where it uses the cellular transcription machinery to ultimately produce the viral progeny. “As reverse transcription happens inside the capsid shell, all the proteins needed to make DNA out of RNA must be inside the capsid. This does not only include viral proteins but also cellular ones,” Garcia-Moreno points out. To test which are the potentially active proteins in infection, the researchers then adapted the method developed in sindbis to study HIV particles. Based on complementary DNA probes that pair with the viral RNA, the bait, the scientists can fish out the HIV RNA together with the proteins that attach to it. They found that 73 cellular proteins are reproducibly packaged with the viral RNA inside the HIV capsid shell. As a proof-of-principle, they removed four of these proteins from human cells using CRISPR/cas9 and discovered they are critical for HIV infection.
Sorting out the molecular mechanisms in HIV infection for new therapies
“After weeding out the proteins needed for infection and replication, we will identify which proteins are incorporated into the virions by active or passive mechanisms and how these proteins regulate infection,” explains Garcia-Moreno. The work has been published in the peer-reviewed articles: ‘Unconventional RNA‐binding proteins step into the virus–host battlefront’(opens in new window) and ‘System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection’(opens in new window). Work in the lab continues and the researchers are actively looking for funding, including support from the MRC and ERC. Castello sums up plans for the future: “We will exploit hRBP-virion results in follow-up collaborations that could lead to patents in the long term for the benefit of patients with HIV.”