Adaptation of Man's making
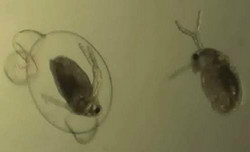
The ADAPT-ENVGENOME(opens in new window) (Environmental adaptation of the genome: A Daphnia model under cultural eutrophication) project has looked at genetic changes in the water flea, Daphnia. This interdisciplinary study integrated methodologies from the fields of genetics, physiology, and resurrection ecology, coupled with competition and juvenile growth rate experiments. The researchers studied the ecosystems in five lakes in Minnesota (U.S.A.) with distinct cultural eutrophication (i.e. man-induced nutrient enrichment) histories. In parallel, the researchers collected genomic data related to gene pathways involved in processing the important element, phosphorus (P), in the water fleas from different time periods. For this purpose, they tapped into the sediment egg banks in these lakes using resurrection ecology methods. The result is a wealth of data on nutrient levels in the environment of the present-day water flea and its relatives across the past several centuries. Data revealed that under competition, Daphnia genotypes (clones) from the lake with more varied eutrophication levels showed competitive advantages at both high and low P feeding supplies. Moreover, this population from the variable lake has higher P retention ability. However, youngsters that do not compete well from Daphnia clones in a lake with virtually constant P supply, exhibited greater plasticity in their growth rate. Most showed a higher growth rate in the early stages of their life cycle. These results were presented at the Evolution Meeting, 2013 in Utah, United States of America. Using filtering procedures from bioinformatics pipelines, 40 000 putative SNP markers were reduced to almost 8 500 highly confident and bi-allelic loci in the water flea genome. Approximately 11 % aligned to unique positions on the reference genome of Daphnia pulex. Significantly, almost 45 % of SNPs in one analysis were found to be involved in lipid, nucleotide and amino acid metabolism and four regulatory pathways. Research results under preparation for a third paper to be submitted for publication indicate that some evolutionary change could be linked to the eutrophication level. Final results should reveal elements of microevolutionary changes under selection pressure of P contamination and researchers will be more likely to find genes involved in these adaptations. Project findings could have a major impact on ecosystem management when considering water pollution, conservation and invasive species.