Understanding plant processes to fight human disease
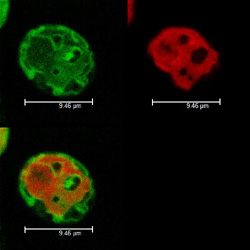
The genetic systems of mitochondria contain a genome that encodes various proteins-coding genes involved in molecular respiration. This is responsible for generating most of the cell's energy and, thus, synthesis of these proteins is critical for survival. This process requires a complete set of transfer ribonucleic acids (tRNAs). But in several organisms, their number cannot adequately ensure mitochondrial translation. To compensate for this insufficiency, nuclear encoded tRNAs have to be imported from the intracellular fluid (cytosol). Although mitochondrial tRNA import is actually a widespread phenomenon and has been experimentally documented even in humans, there is still an incomplete understanding of the relevant mechanisms. The 'Dissecting the cytosolic tRNA import process in mitochondria of the unicellular green alga Chlamydomonas reinhardtii' (CHLAMITitRNA) project set out to improve on this and learn more about regulation of the tRNA import process and its machinery. The green algae Chlamydomonas reinhardtii was selected as the best-suited model for this study. Regarding the import process, members of this EU-funded project observed that the mitochondrial population of nuclear-encoded tRNAs is primarily complementary to those encoded in the mitochondrial genome. Also, experimental results and data analysis suggested that the identity and the levels of imported tRNAs can be defined by the information contained in the mitochondrial genome. To test this, CHLAMITRNA researchers modified the Chlamydomonas mitochondrial genome in order to observe how it would affect tRNA import. Experimental results were not able to prove the hypothesis. RNA silencing was used to investigate the Voltage voltage-Dependent dependent anion channel (VDAC) — a major component of tRNA through the outer mitochondrial membrane. This approach succeeded in downregulating the expression of VDACI and VDACII proteins showing that physiological activities such as respiration can be affected in mutations. Enhanced understanding of the import of mitochondrial nucleic acids has important technological applications. It holds promise for developing strategies to address cases where mitochondrial transformation is still difficult or even impossible. This is turn could offer knowledge necessary to develop strategies for therapeutic intervention in human genetic diseases caused by mutations in mitochondrial tRNA genes.