Printing peptides
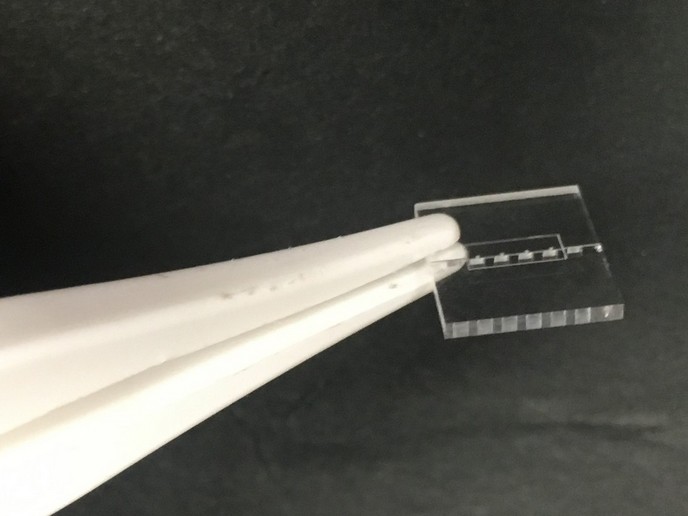
Proteins are central to virtually all biological functions They are typically made up of very long chains of peptides (themselves made from combinations of amino acids), folded into complex three-dimensional (3D) structures. The use of peptides to probe protein structure and function as well as for therapies is becoming more and more popular. Combinatorial chemistry refers to the synthesis of chemical compounds as libraries of many molecules, and the screening of those libraries for candidates with certain qualities. The goal of the EU-funded 'Combinatorial synthesis of peptide arrays with a laser printer' (Peplaser) project was to dramatically increase the number of peptides synthesised by small devices for high-throughput analysis. Peplaser used an amino acid particle-based method for combinatorial synthesis of complex peptide arrays. Solid amino acid particles contain activated chemical building blocks or amino acid derivatives. Peplaser developed a system to deliver the particles to a modified colour laser printer that produces amino acid derivatives. Repeated cycles of printing and 'melting' yielded the peptide arrays. Scientists combined this technology with bioinformatics and readout tools to create small and robust machines. The peptide laser printer reliably synthesised 275 000 high-quality peptides in a small square of 20 cm by 20 cm. It did so at a cost estimated to be 1 000 times less per peptide than that of the current state of the art. During the course of the project, scientists were able to increase the machine’s synthesis capacity from 0,5 million peptides per month to more than 10 million. Peplaser technology was used to find and analyse antibiotic peptides and to correlate peptide patterns with prognoses in tumour patients. It was also used to diagnose infection thanks to the presence of antibodies against a certain bacteria. The technology is currently commercially available from one of the project partners, which is a small and medium-sized enterprise (SME). The Peplaser project outcomes are expected to revolutionise the field of proteomics much as the high-density, oligonucleotide array-based lithographic technologies did for genomics. Rapid, accurate and high-throughput analyses, reminiscent of the mathematical calculations done by a computer rather than by hand, open new doors to problem solving in this field.