How the body fights infection can provide clues to stomach cancer risk
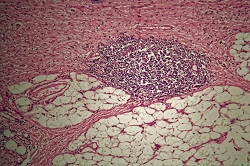
EU-funded scientists working on the HELICOMARK project have found ways to assess stomach cancer risk in blood samples using the body’s immune response as a tool. Around 700 000 people die each year around the world from stomach cancer, the third most common cause of cancer death. Together Japan, Korea and China account for almost 70 % of all cases but it is also common in Latin America and Eastern Europe, and patients generally have a poor survival prognosis. A major cause of stomach cancer is the Helicobacter pylori germ. “H. pylori as a bacterium is very genetically variable and the variation more or less follows ethnicity, with East Asian versions of H. pylori being more carcinogenic,” says project coordinator Professor Samuel Lundin, a professor in the department of Microbiology and Immunology at the University of Gothenburg, Sweden. “We have been analysing in very high detail the antibody responses to H. pylori to pinpoint the more risky H. pylori bacteria present in blood samples, and analysing how this is associated with stomach cancer risk.” Peptide arrays Researchers produce peptide arrays to profile a patient’s immune response to H. pylori by synthesising peptides on a glass chip. “Technologies are available that allow us to add one amino acid letting it react, then washing it away and adding another amino acid till a peptide sequence is produced on the glass chip,” Prof Lundin explains. Serum samples from Nicaragua, where there is a high risk of stomach cancer, and Sweden, were added to the 200 000 peptides on the chip. “Depending on the antibodies produced, some of them will bind to some of the peptides. Then you add another antibody that is fluorescently labelled so you light up those peptides,” Prof Lundin says. “The assay shows the existence of these antibodies, so what we are doing is mapping the antibodies by using the peptide chip,” he explains. “From this we get a score of how well the antibodies have bound to peptides, and to which of the 200 000 peptides.” This produces a specific ‘fingerprint’ for each person which can then be compared with those who developed the cancer. Mapping proteins “We mapped all the proteins that will elicit an antibody response if you have H. pylori, and all the epitopes of those proteins (the parts of the proteins to which antibodies bind),” says Professor Lundin. “We have shown that only two or three of those epitopes are diagnostic in patients, the others produce antibodies even if the patient doesn’t have H. pylori.” This method of mapping the epitopes has been patented. “We identified the peptides, where, if the patient has antibodies to them, we know there is a high risk of developing stomach cancer,” Professor Lundin explains. The key to the project’s success was access to blood samples as well as biopsies from the same person. “We can culture the bacteria from the biopsies and analyse the person’s H. pylori strains using genomic analysis,” he continues. The same person’s antibody response is analysed to see the peptide sequences they respond to. “It’s uncommon to have all of this information together and that was crucial for us.” Professor Lundin received a three-year Marie Curie grant that funded two years in Perth, Australia, working with Nobel Laureate Barry Marshall, one of the two scientists who discovered H. pylori.