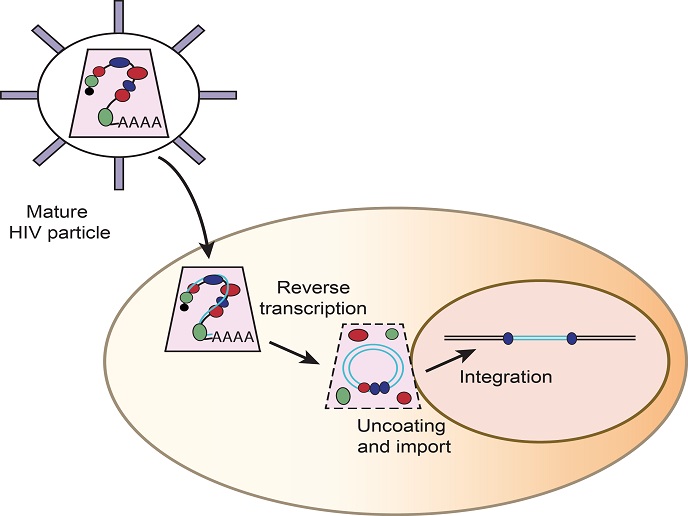
Silencing nuclear RNAs
The aim of the EU-funded project NONCODOWN (Optimizing antisense oligonucleotides for efficient and specific knockdown of nuclear non-coding RNA) was to develop the best technology for deactivating (silencing) nuclear RNAs. Scientists applied the most successful techniques from the field of nucleic acid chemistry. They were based on locked nucleic acid (LNA), also known as inaccessible RNA, and antisense oligonucleatides (ASOs), known as gapmers. They also developed new designs of peptide nucleic acid (PNA)-DNA-LNA chimeric oligomers for degrading structured RNA targets. A series of optimised ASOs were developed for targeting and degrading structured RNAs. This was achieved by containing a partial sequence of PNA and part of DNA, joined by a conformationally locked nucleotide. Chemical designs were compared for silencing a nuclear-retained RNA transcript and included silencing RNAs (siRNAs), single-strand siRNAs and LNA-modified ASOs. Researchers found that the ASOs were the most effective in carrying out this task. NONCODOWN also screened chemically modified oligonucleotides such as LNA and PNA for their ability to block the action of a bacterial regulatory non coding ncRNA. They were also used to identify potent inhibitors.