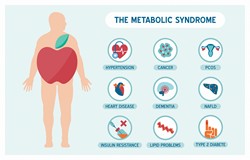
Molecular determinants in metabolic syndrome
Apolipoprotein E (APOE) associates with lipoproteins and mediates their clearance. In humans, the ApoE4 allele is linked with an increased risk to develop cardiovascular disease but the underlying mechanism is not clear. The scope of the EU-funded APOMET (Apolipoprotein E gene in the metabolic syndrome) project was to investigate the role of APOE in MetS. In this context, they employed a multidisciplinary approach that combined in vivo analysis of transgenic mice with epidemiological analysis of human populations. Researchers based their epidemiological analysis on the Aragon Workers Health Study (AWHS) cohort, an ongoing study targeted to study genetic determinants and lifestyle habits in cardiovascular disease. Using this cohort, they discovered an association between APOE2 and an increased body mass index, as well as a dose-dependent association between the ApoE4 allele and an increased risk for MetS. To elucidate the molecular mechanisms underlying APOE-directed metabolic alterations, scientists worked under the hypothesis that APOE4 drove a whole-body metabolic shift toward increased lipid oxidation. Towards this goal, they utilised humanised mice where the endogenous ApoE gene was replaced with either ApoE3 or ApoE4 alleles. Results indicated that mice carrying the ApoE4 allele used lipids rather than carbohydrates as a fuel source. They also presented with increased body temperature, augmented cold tolerance and more metabolically active brown adipose tissue compared with mice carrying the ApoE3 allele. These results suggested that ApoE4 mice might resist weight gain via an APOE4-directed global metabolic shift toward lipid oxidation. Taken together, the findings of the APOMET study established an association between APOE and metabolism. This represented a critical first step in the development of APOE-directed therapies for a large percentage of the population affected by various metabolic disorders.