Protein complexes, chromatin and disease
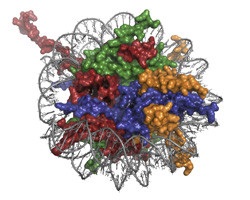
Chromatin, a macromolecular DNA–protein complex, participates in the regulation of processes of DNA metabolism, such as replication, transcription and repair. These processes are controlled by protein complexes that interact with nucleosomes, the structural sub-units of chromatin. The project MSL1-MSL3-MOF was dedicated to research on one such regulatory complex. The MOF protein is an acetyl transferase that participates in DNA transcription. The other two proteins in the complex (MSL1 and MSL3) enhance the enzymatic activity of MOF. The details of interaction between the components of this complex with each other and with chromatin are unknown — a knowledge gap this study sought to fill. Researchers reconstituted, purified and biochemically characterised the protein complex, and mapped the interactions between the components of the complex to define its catalytic activity. The team performed a preliminary 3D reconstruction of the regulatory complex, and designed and tested a protocol for the assembly and purification of chromatin substrate. Prior to attempting X-ray crystallography of MSL1-MSL3-MOF, it was necessary to define the conditions for crystallisation and structural analysis using a model system. As crystallography of large DNA–protein complexes is very challenging, a well-characterised nucleosome-binding protein (bromo-adjacent homology (BAH) domain) was used to define the right conditions. The structure of BAH was successfully solved and revealed a conformational change induced by N-terminal acetylation, which is a common post-translational modification in eukaryotes. The found conformational change positioned acetylated BAH closer to the nucleosome compared to non-acetylated BAH. This result suggests a general role of N-acetylation in protein interactions. Thus, using this model system allowed not only to define the conditions for X-ray crystallography of the MOF complex, but also revealed novel structural information. As many protein complexes regulating chromatin are linked to disease, resolving their structure will enable design of specific drugs to address the problem. The work performed in this project was a first step towards this important goal.