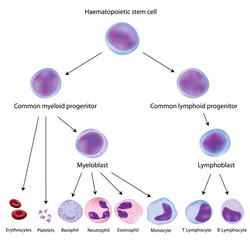
The molecular determinants of haematopoiesis
Transcription factors are critical for normal haematopoietic differentiation during development and homeostasis. Reduction in the levels of the transcription factor PU.1 leads to the development of acute myeloid leukaemia (AML). Evidence indicates that PU.1 expression is regulated at the chromatin level. However, little is known regarding how the transcriptionally active or silent state of chromatin is propagated and maintained. Scientists on the EU-funded PU.1 ANTISENSE (Role of antisense RNAs on PU.1 regulation and hematopoiesis) project set out to investigate the role of non-coding antisense transcripts in PU.1 gene regulation. Their work focused on the interplay among these non-coding RNAs and known epigenetic marks. As a first step, scientists quantified the levels of PU.1 and its non-coding transcripts during blood cell differentiation, alongside the methylation status of the PU.1 gene promoter. They observed a strong inverse correlation of PU.1 mRNA and antisense transcription in different blood cell types. Ablation of non-coding antisense transcripts altered chromatin structure and induced PU.1 expression and myeloid cell differentiation. Similar observations were made following the use of antisense transcripts in animal models of leukaemia: leukaemic cells underwent apoptosis and mice survived longer. Furthermore, they discovered a novel mechanism whereby oncogenic fusion proteins that result from chromosomal translocations induce antisense transcription and promoter methylation. Collectively, the consortium demonstrated that antisense technology could be utilised therapeutically to change the fate of leukaemic cells. They also changed our view on how chromosomal translocation products promote leukaemogenesis.