The mathematics of genome evolution
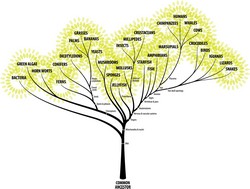
The notion that all forms of life are genetically related is one of the most romantic notions in science. This so-called phylogeny of organisms implies that different species arise from previous forms by descent. In addition, all organisms are connected by the passage of genes along the branches of the tree of life. The leaves of this vast evolutionary tree correspond to organisms that are alive today. The roots represent the last common ancestors of all species in the tree. Researchers assume that genetic information, including DNA and proteins, evolved from the roots to leaves in accordance with general Markov models. Within the project TREEMODELS (Algebraic statistics of general Markov models), the scientists studied general Markov models. This model class contains many simulations used in phylogenetics to explain similarities among plants, animals and microorganisms. These statistical models are algebraic in that they can be defined in terms of polynomial constraints or parameterisations. Analysis of general Markov models was based on the use of algebraic geometry that enables representation of independencies among various variables. Algebraic statistics offered a computational framework to address problems such as label switching that make the interpretation of results difficult. The TREEMODELS team also proposed alternative, simpler statistical models of processes that generate data from genomes and, in what is known as statistical inference, drew conclusions about these processes. Genome sequences are the blueprint for life, and yet their function and evolution are poorly understood. These new models, called marginal supermodels, proved to represent biological evolution more efficiently than standard phylogenetic tree models. The team ensured that the tree parameters were identifiable so that evolutionary histories can be consistently inferred. Scientists shed further insight into the geometry of the different tree models by studying different algebraic varieties. Using tree cumulants, they proved that the secant variety of the Segre variety is toric. Focus was also placed on the strong positivity and convexity properties of another projective algebraic variety known as exponential variety. Algebraic statistics is a new field whose scope is still widening. The TREEMODELS project took steps along the algebraic statistics path with the aim of developing inference tools necessary for biological sequence analysis. A rigorous computational framework is expected to emerge for the organisation of biological knowledge.