Insight into the role of bacterial ecosystems in the scourge of antibiotic resistance development
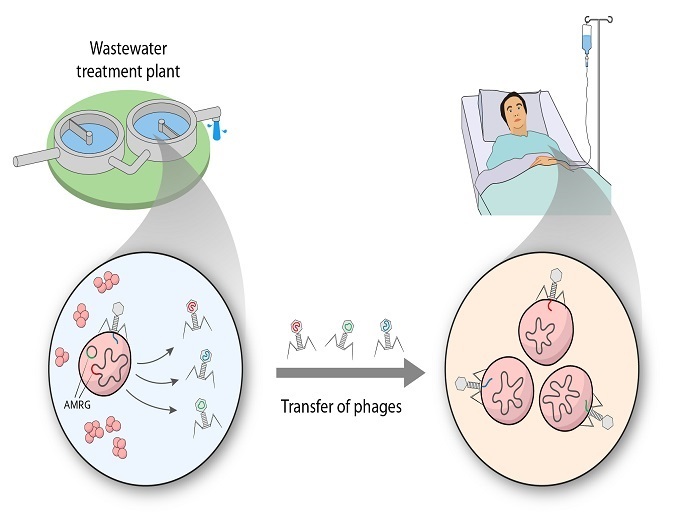
Bacteriophages are the most abundant organisms on Earth and play a central role in microbial physiology and ecosystem homeostasis. They are species specific and do not impact human commensal flora, such as gut bacteria, rendering them suitable as a promising alternative to antibiotics for killing pathogenic bacteria. However, recent data reveal that phages may have a broader host range than traditionally thought and indirect evidence suggests that bacteriophages may be implicated in the transmission of AMR genes.
Investigation of bacteriophages in human-affected environments
The ROPHARE project was undertaken with the support of the Marie Skłodowska-Curie (MSC) programme and aimed to investigate the involvement of bacteriophages in the horizontal gene transfer of AMR genes among bacteria. Researchers worked under the theory that the global rise in AMR cannot be explained purely on the basis of inappropriate antibiotics prescription. “There is increasing consensus that microbial communities, antibiotics and AMR genes from different ecosystems cooperate to disseminate AMR genes and propagate resistance,” explains the MSC research fellow Elena Gómez-Sanz. To study this complex interaction, the researchers studied two environmental systems managed by man – agricultural soil and a wastewater treatment plant (WWTP). They isolated and characterised approximately 100 novel staphylococcal bacteriophages in WWTP water (untreated and treated) and extracted the genetic material of microbial communities from the agricultural soil. This enabled them to unveil hundreds of novel environmental bacteriophages and determine if bacteriophages carry AMR genes and how they mediate their spread to the entire community. Results also revealed that several staphylococcal strains can transfer, although at low rates, specific resistance genes to clinically relevant species, highlighting the importance of the staphylococcal bacteriophage-bacterial network. This took place through plasmids, small circular pieces of DNA separate from the main bacterial chromosome. Following sequencing and extensive bioinformatics analysis, scientists identified an unprecedented number of novel bacteriophages as well as new AMR genes. Most identified bacteriophages exhibited a broad host-range, being able to infect clinically relevant multidrug-resistant strains, and some carried AMR genes of clinical relevance. Annotation of the newly identified AMR genes will advance existing knowledge on the genetics of resistance.
ROPHARE significance and future prospects
ROPHARE provided unprecedented information on microbial ecology and composition of agricultural soils, contributing to the understanding of the role of bacteriophages on ecology and microorganism evolution. Significantly, the diversity they discovered of both viruses and bacteria indicated that agricultural fertiliser application modifies microbial composition. By focusing on the dynamics of man-managed systems, ROPHARE will contribute to a better understanding on how human intervention affects the transmission of AMR genes to animal and human microbiomes. These results can also point towards potential anti-microbial agents or antibiotic combinations capable of restraining AMR spread. From a therapeutic perspective, the identification and characterisation of novel bacteriophages can pave the way for the development of alternative anti-microbial treatments. “The molecular analysis of the identified bacteriophages has allowed us to determine the most promising candidates for anti-microbial strategies with little AMR gene mobilisation capacity,” emphasises Gómez-Sanz. Currently, approximately 700 000 people die of AMR worldwide, a number that is projected to increase exponentially by 2050. ROPHARE results have the potential to help stakeholders formulate the necessary risk mitigation strategies. Knowing the potential reservoirs of AMR genes in the environment will help make decisions on the use of animal waste fertilisers in soil systems, wastewater management and other strategies to avoid AMR spread.